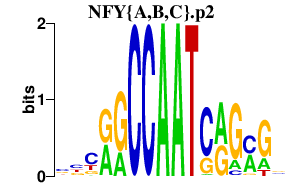
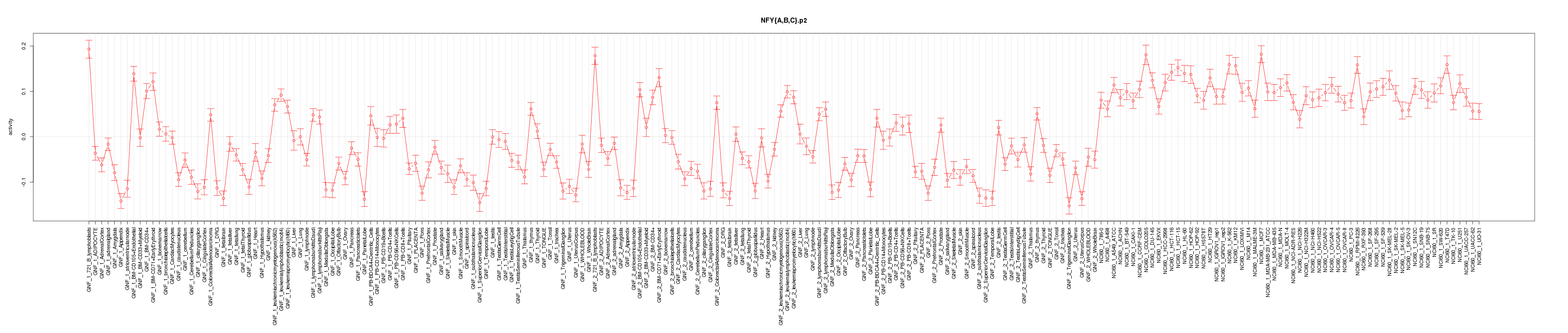
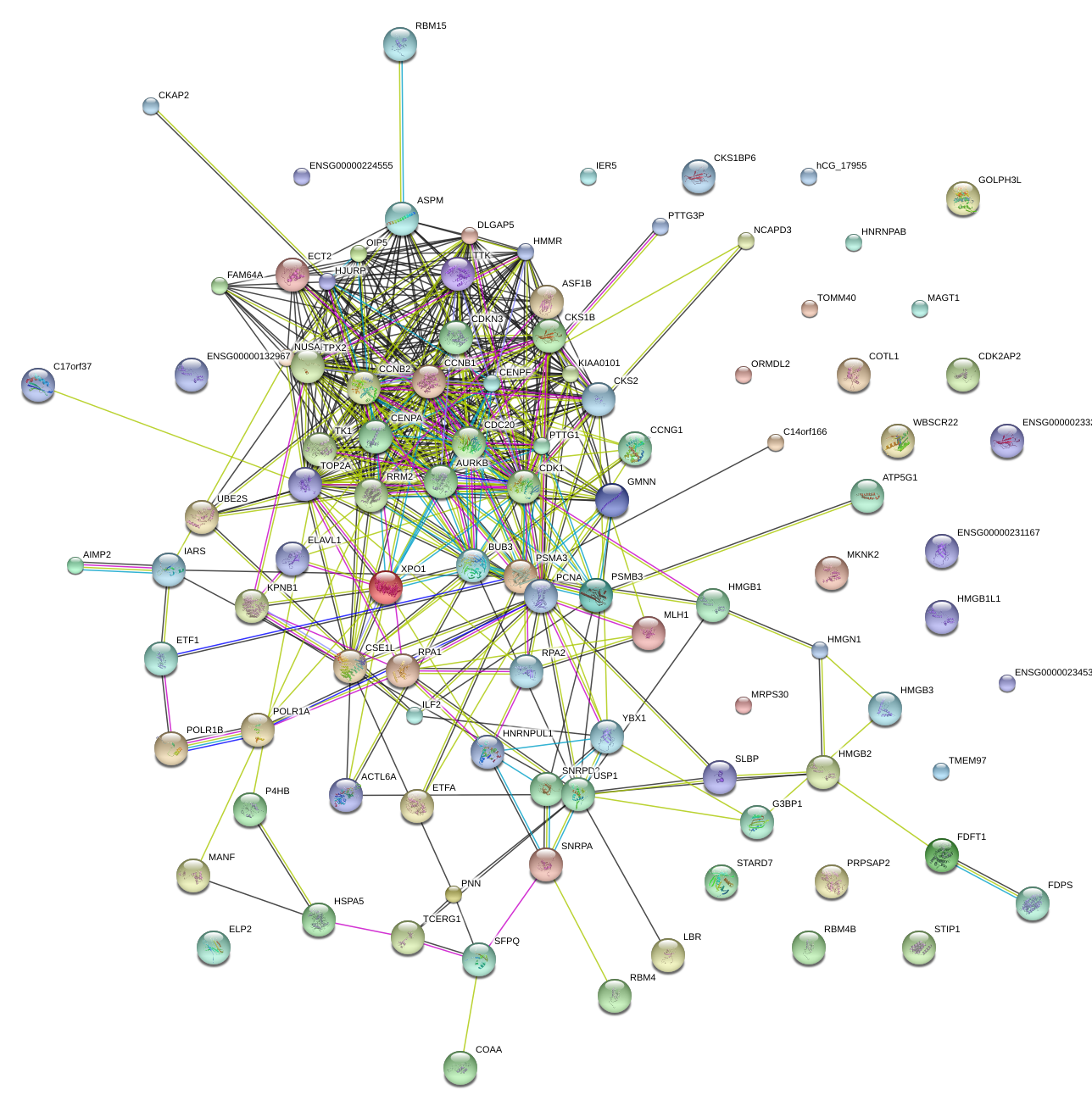
|
1.28
|
2.00e-37
|
GO:0009987
|
cellular process
|
|
1.55
|
8.59e-37
|
GO:0044237
|
cellular metabolic process
|
|
3.41
|
2.27e-36
|
GO:0022403
|
cell cycle phase
|
|
3.53
|
3.33e-33
|
GO:0000278
|
mitotic cell cycle
|
|
3.06
|
6.92e-33
|
GO:0022402
|
cell cycle process
|
|
2.77
|
2.04e-32
|
GO:0007049
|
cell cycle
|
|
1.66
|
1.81e-30
|
GO:0044260
|
cellular macromolecule metabolic process
|
|
1.43
|
1.40e-28
|
GO:0008152
|
metabolic process
|
|
3.75
|
2.26e-28
|
GO:0000279
|
M phase
|
|
1.47
|
2.04e-27
|
GO:0044238
|
primary metabolic process
|
|
1.98
|
6.22e-26
|
GO:0090304
|
nucleic acid metabolic process
|
|
2.19
|
8.55e-26
|
GO:0006996
|
organelle organization
|
|
1.77
|
1.16e-25
|
GO:0034641
|
cellular nitrogen compound metabolic process
|
|
1.75
|
2.61e-25
|
GO:0006807
|
nitrogen compound metabolic process
|
|
1.83
|
6.93e-25
|
GO:0006139
|
nucleobase, nucleoside, nucleotide and nucleic acid metabolic process
|
|
4.09
|
9.06e-24
|
GO:0000087
|
M phase of mitotic cell cycle
|
|
1.53
|
1.88e-23
|
GO:0043170
|
macromolecule metabolic process
|
|
3.65
|
5.46e-21
|
GO:0010564
|
regulation of cell cycle process
|
|
1.78
|
1.10e-20
|
GO:0071842
|
cellular component organization at cellular level
|
|
1.76
|
1.33e-20
|
GO:0071841
|
cellular component organization or biogenesis at cellular level
|
|
3.87
|
3.21e-20
|
GO:0000280
|
nuclear division
|
|
3.87
|
3.21e-20
|
GO:0007067
|
mitosis
|
|
3.75
|
2.46e-19
|
GO:0048285
|
organelle fission
|
|
2.79
|
1.18e-18
|
GO:0051276
|
chromosome organization
|
|
1.62
|
7.61e-18
|
GO:0016043
|
cellular component organization
|
|
1.74
|
1.11e-17
|
GO:0009058
|
biosynthetic process
|
|
3.59
|
1.34e-17
|
GO:0051325
|
interphase
|
|
1.60
|
1.37e-17
|
GO:0071840
|
cellular component organization or biogenesis
|
|
3.60
|
2.23e-17
|
GO:0051329
|
interphase of mitotic cell cycle
|
|
4.06
|
3.99e-17
|
GO:0071156
|
regulation of cell cycle arrest
|
|
2.69
|
4.60e-17
|
GO:0051726
|
regulation of cell cycle
|
|
3.33
|
1.03e-16
|
GO:0051301
|
cell division
|
|
4.07
|
1.65e-16
|
GO:0000075
|
cell cycle checkpoint
|
|
1.71
|
3.76e-15
|
GO:0044249
|
cellular biosynthetic process
|
|
2.76
|
4.67e-14
|
GO:0034621
|
cellular macromolecular complex subunit organization
|
|
2.28
|
9.75e-14
|
GO:0043933
|
macromolecular complex subunit organization
|
|
2.57
|
9.81e-14
|
GO:0006259
|
DNA metabolic process
|
|
2.27
|
1.60e-13
|
GO:0033554
|
cellular response to stress
|
|
4.29
|
5.44e-13
|
GO:0071103
|
DNA conformation change
|
|
4.99
|
9.68e-13
|
GO:0065004
|
protein-DNA complex assembly
|
|
4.86
|
9.70e-13
|
GO:0071824
|
protein-DNA complex subunit organization
|
|
2.96
|
2.05e-12
|
GO:0034622
|
cellular macromolecular complex assembly
|
|
4.52
|
2.14e-12
|
GO:0006323
|
DNA packaging
|
|
2.34
|
3.40e-12
|
GO:0065003
|
macromolecular complex assembly
|
|
1.79
|
5.26e-12
|
GO:0009059
|
macromolecule biosynthetic process
|
|
1.78
|
1.51e-11
|
GO:0034645
|
cellular macromolecule biosynthetic process
|
|
1.76
|
1.52e-11
|
GO:0016070
|
RNA metabolic process
|
|
4.03
|
6.08e-10
|
GO:0006333
|
chromatin assembly or disassembly
|
|
1.89
|
7.26e-10
|
GO:0044085
|
cellular component biogenesis
|
|
4.67
|
8.73e-10
|
GO:0031497
|
chromatin assembly
|
|
2.50
|
1.18e-09
|
GO:0006325
|
chromatin organization
|
|
4.76
|
1.25e-09
|
GO:0006334
|
nucleosome assembly
|
|
1.56
|
1.82e-09
|
GO:0044267
|
cellular protein metabolic process
|
|
3.92
|
3.20e-09
|
GO:0006310
|
DNA recombination
|
|
4.44
|
3.77e-09
|
GO:0034728
|
nucleosome organization
|
|
4.40
|
4.99e-09
|
GO:0007059
|
chromosome segregation
|
|
2.37
|
5.32e-09
|
GO:0006974
|
response to DNA damage stimulus
|
|
1.91
|
5.92e-09
|
GO:0022607
|
cellular component assembly
|
|
3.63
|
8.92e-09
|
GO:0010498
|
proteasomal protein catabolic process
|
|
3.63
|
8.92e-09
|
GO:0043161
|
proteasomal ubiquitin-dependent protein catabolic process
|
|
1.65
|
1.03e-08
|
GO:0010467
|
gene expression
|
|
7.16
|
1.13e-08
|
GO:0016126
|
sterol biosynthetic process
|
|
4.73
|
2.34e-08
|
GO:0000236
|
mitotic prometaphase
|
|
2.29
|
3.08e-08
|
GO:0016071
|
mRNA metabolic process
|
|
2.91
|
3.34e-08
|
GO:0007017
|
microtubule-based process
|
|
7.55
|
6.18e-08
|
GO:0006695
|
cholesterol biosynthetic process
|
|
2.61
|
9.33e-08
|
GO:0006281
|
DNA repair
|
|
2.03
|
1.12e-07
|
GO:0071844
|
cellular component assembly at cellular level
|
|
2.25
|
1.32e-07
|
GO:0044265
|
cellular macromolecule catabolic process
|
|
3.59
|
1.83e-07
|
GO:0000082
|
G1/S transition of mitotic cell cycle
|
|
2.81
|
2.06e-07
|
GO:0007346
|
regulation of mitotic cell cycle
|
|
1.75
|
2.21e-07
|
GO:0051641
|
cellular localization
|
|
2.13
|
2.37e-07
|
GO:0009057
|
macromolecule catabolic process
|
|
1.60
|
3.42e-07
|
GO:0044281
|
small molecule metabolic process
|
|
3.95
|
4.42e-07
|
GO:0000086
|
G2/M transition of mitotic cell cycle
|
|
3.19
|
4.82e-07
|
GO:0006260
|
DNA replication
|
|
4.45
|
1.42e-06
|
GO:0000216
|
M/G1 transition of mitotic cell cycle
|
|
1.74
|
2.45e-06
|
GO:0044248
|
cellular catabolic process
|
|
2.03
|
3.55e-06
|
GO:0006396
|
RNA processing
|
|
2.55
|
3.65e-06
|
GO:0008380
|
RNA splicing
|
|
3.53
|
3.73e-06
|
GO:0007093
|
mitotic cell cycle checkpoint
|
|
2.93
|
6.35e-06
|
GO:0006457
|
protein folding
|
|
1.46
|
8.66e-06
|
GO:0006950
|
response to stress
|
|
1.63
|
9.32e-06
|
GO:0009056
|
catabolic process
|
|
1.72
|
1.20e-05
|
GO:0051649
|
establishment of localization in cell
|
|
3.19
|
1.50e-05
|
GO:0000226
|
microtubule cytoskeleton organization
|
|
2.03
|
1.52e-05
|
GO:0071822
|
protein complex subunit organization
|
|
4.10
|
1.84e-05
|
GO:0007088
|
regulation of mitosis
|
|
4.10
|
1.84e-05
|
GO:0051783
|
regulation of nuclear division
|
|
5.49
|
2.01e-05
|
GO:0000819
|
sister chromatid segregation
|
|
3.57
|
2.05e-05
|
GO:0051320
|
S phase
|
|
2.82
|
2.95e-05
|
GO:0000375
|
RNA splicing, via transesterification reactions
|
|
2.24
|
4.14e-05
|
GO:0006091
|
generation of precursor metabolites and energy
|
|
3.50
|
6.18e-05
|
GO:0000084
|
S phase of mitotic cell cycle
|
|
1.83
|
7.65e-05
|
GO:0046907
|
intracellular transport
|
|
3.97
|
7.73e-05
|
GO:0031145
|
anaphase-promoting complex-dependent proteasomal ubiquitin-dependent protein catabolic process
|
|
1.37
|
8.49e-05
|
GO:0019538
|
protein metabolic process
|
|
2.36
|
1.04e-04
|
GO:0006511
|
ubiquitin-dependent protein catabolic process
|
|
4.93
|
1.18e-04
|
GO:0007051
|
spindle organization
|
|
2.19
|
1.54e-04
|
GO:0006397
|
mRNA processing
|
|
2.30
|
1.62e-04
|
GO:0051603
|
proteolysis involved in cellular protein catabolic process
|
|
3.55
|
1.69e-04
|
GO:0006694
|
steroid biosynthetic process
|
|
2.31
|
1.93e-04
|
GO:0019941
|
modification-dependent protein catabolic process
|
|
2.29
|
2.36e-04
|
GO:0043632
|
modification-dependent macromolecule catabolic process
|
|
2.25
|
2.93e-04
|
GO:0044257
|
cellular protein catabolic process
|
|
2.20
|
3.57e-04
|
GO:0030163
|
protein catabolic process
|
|
5.16
|
4.31e-04
|
GO:0000070
|
mitotic sister chromatid segregation
|
|
2.65
|
5.29e-04
|
GO:0000377
|
RNA splicing, via transesterification reactions with bulged adenosine as nucleophile
|
|
2.65
|
5.29e-04
|
GO:0000398
|
nuclear mRNA splicing, via spliceosome
|
|
3.21
|
6.60e-04
|
GO:0051640
|
organelle localization
|
|
1.42
|
9.96e-04
|
GO:0048523
|
negative regulation of cellular process
|
|
10.57
|
1.29e-03
|
GO:0000085
|
G2 phase of mitotic cell cycle
|
|
10.57
|
1.29e-03
|
GO:0051319
|
G2 phase
|
|
3.19
|
1.31e-03
|
GO:0007126
|
meiosis
|
|
3.19
|
1.31e-03
|
GO:0051327
|
M phase of meiotic cell cycle
|
|
1.96
|
1.51e-03
|
GO:0070271
|
protein complex biogenesis
|
|
3.16
|
1.56e-03
|
GO:0051321
|
meiotic cell cycle
|
|
5.93
|
1.78e-03
|
GO:0006271
|
DNA strand elongation involved in DNA replication
|
|
1.98
|
1.90e-03
|
GO:0006066
|
alcohol metabolic process
|
|
2.00
|
1.99e-03
|
GO:0044262
|
cellular carbohydrate metabolic process
|
|
3.02
|
2.11e-03
|
GO:0010639
|
negative regulation of organelle organization
|
|
1.94
|
2.66e-03
|
GO:0006461
|
protein complex assembly
|
|
3.50
|
2.82e-03
|
GO:0051656
|
establishment of organelle localization
|
|
1.37
|
3.21e-03
|
GO:0048522
|
positive regulation of cellular process
|
|
5.54
|
4.02e-03
|
GO:0022616
|
DNA strand elongation
|
|
1.42
|
4.04e-03
|
GO:0043412
|
macromolecule modification
|
|
3.17
|
4.39e-03
|
GO:0016125
|
sterol metabolic process
|
|
1.79
|
5.05e-03
|
GO:0005975
|
carbohydrate metabolic process
|
|
3.21
|
6.20e-03
|
GO:0051340
|
regulation of ligase activity
|
|
1.89
|
6.63e-03
|
GO:0007010
|
cytoskeleton organization
|
|
1.17
|
7.16e-03
|
GO:0050794
|
regulation of cellular process
|
|
2.74
|
8.43e-03
|
GO:0051052
|
regulation of DNA metabolic process
|
|
2.10
|
8.79e-03
|
GO:0033043
|
regulation of organelle organization
|
|
1.36
|
9.19e-03
|
GO:0048519
|
negative regulation of biological process
|
|
2.94
|
9.49e-03
|
GO:0031570
|
DNA integrity checkpoint
|
|
1.15
|
9.63e-03
|
GO:0065007
|
biological regulation
|
|
3.66
|
1.04e-02
|
GO:0010948
|
negative regulation of cell cycle process
|
|
1.41
|
1.04e-02
|
GO:0006464
|
protein modification process
|
|
2.02
|
1.11e-02
|
GO:0016568
|
chromatin modification
|
|
5.04
|
1.18e-02
|
GO:0051297
|
centrosome organization
|
|
3.32
|
1.22e-02
|
GO:0051351
|
positive regulation of ligase activity
|
|
3.19
|
1.24e-02
|
GO:0051438
|
regulation of ubiquitin-protein ligase activity
|
|
4.27
|
1.32e-02
|
GO:0006458
|
'de novo' protein folding
|
|
1.59
|
1.34e-02
|
GO:0032774
|
RNA biosynthetic process
|
|
1.27
|
1.34e-02
|
GO:0080090
|
regulation of primary metabolic process
|
|
2.18
|
1.35e-02
|
GO:0015980
|
energy derivation by oxidation of organic compounds
|
|
10.07
|
1.38e-02
|
GO:0002566
|
somatic diversification of immune receptors via somatic mutation
|
|
10.07
|
1.38e-02
|
GO:0016446
|
somatic hypermutation of immunoglobulin genes
|
|
1.27
|
1.43e-02
|
GO:0031323
|
regulation of cellular metabolic process
|
|
1.55
|
1.60e-02
|
GO:0008104
|
protein localization
|
|
1.25
|
1.75e-02
|
GO:0019222
|
regulation of metabolic process
|
|
1.56
|
1.81e-02
|
GO:0032268
|
regulation of cellular protein metabolic process
|
|
1.55
|
1.84e-02
|
GO:0010941
|
regulation of cell death
|
|
1.56
|
1.96e-02
|
GO:0042981
|
regulation of apoptosis
|
|
3.34
|
2.04e-02
|
GO:0051439
|
regulation of ubiquitin-protein ligase activity involved in mitotic cell cycle
|
|
2.74
|
2.06e-02
|
GO:0046034
|
ATP metabolic process
|
|
1.28
|
2.06e-02
|
GO:0060255
|
regulation of macromolecule metabolic process
|
|
1.15
|
2.07e-02
|
GO:0050789
|
regulation of biological process
|
|
3.29
|
2.46e-02
|
GO:0051443
|
positive regulation of ubiquitin-protein ligase activity
|
|
3.16
|
2.47e-02
|
GO:0018196
|
peptidyl-asparagine modification
|
|
3.16
|
2.47e-02
|
GO:0018279
|
protein N-linked glycosylation via asparagine
|
|
1.87
|
2.56e-02
|
GO:0008283
|
cell proliferation
|
|
1.53
|
2.68e-02
|
GO:0051246
|
regulation of protein metabolic process
|
|
1.59
|
2.75e-02
|
GO:0055114
|
oxidation-reduction process
|
|
1.54
|
2.78e-02
|
GO:0043067
|
regulation of programmed cell death
|
|
1.50
|
2.83e-02
|
GO:0050790
|
regulation of catalytic activity
|
|
5.04
|
3.20e-02
|
GO:0071174
|
mitotic cell cycle spindle checkpoint
|
|
3.71
|
3.47e-02
|
GO:0000910
|
cytokinesis
|
|
1.47
|
3.60e-02
|
GO:0033036
|
macromolecule localization
|
|
2.88
|
3.62e-02
|
GO:0090068
|
positive regulation of cell cycle process
|
|
4.49
|
4.09e-02
|
GO:0031023
|
microtubule organizing center organization
|
|
2.85
|
4.19e-02
|
GO:0000077
|
DNA damage checkpoint
|
|
5.44
|
4.28e-02
|
GO:0002200
|
somatic diversification of immune receptors
|
|
1.30
|
4.54e-02
|
GO:0048518
|
positive regulation of biological process
|
|
2.82
|
4.83e-02
|
GO:0042770
|
signal transduction in response to DNA damage
|